Overview
INSIDe will develop a new platform for integrative data-driven analysis of the spread of infectious diseases. To this end, the consortium brings together leading experts in mathematical and computational modelling, epidemiology and infectious disease research. INSIDe will combine and further develop approaches developed independently by the partners to allow flexible integration of different data sources, including (i) official case numbers, mortality, vaccination and hospitalisation statistics, (ii) representative cohort studies and (iii) wastewater samples. The use of multi-resolution simulation models and comprehensive observational process models will enable in-depth assessment and study of large regions.
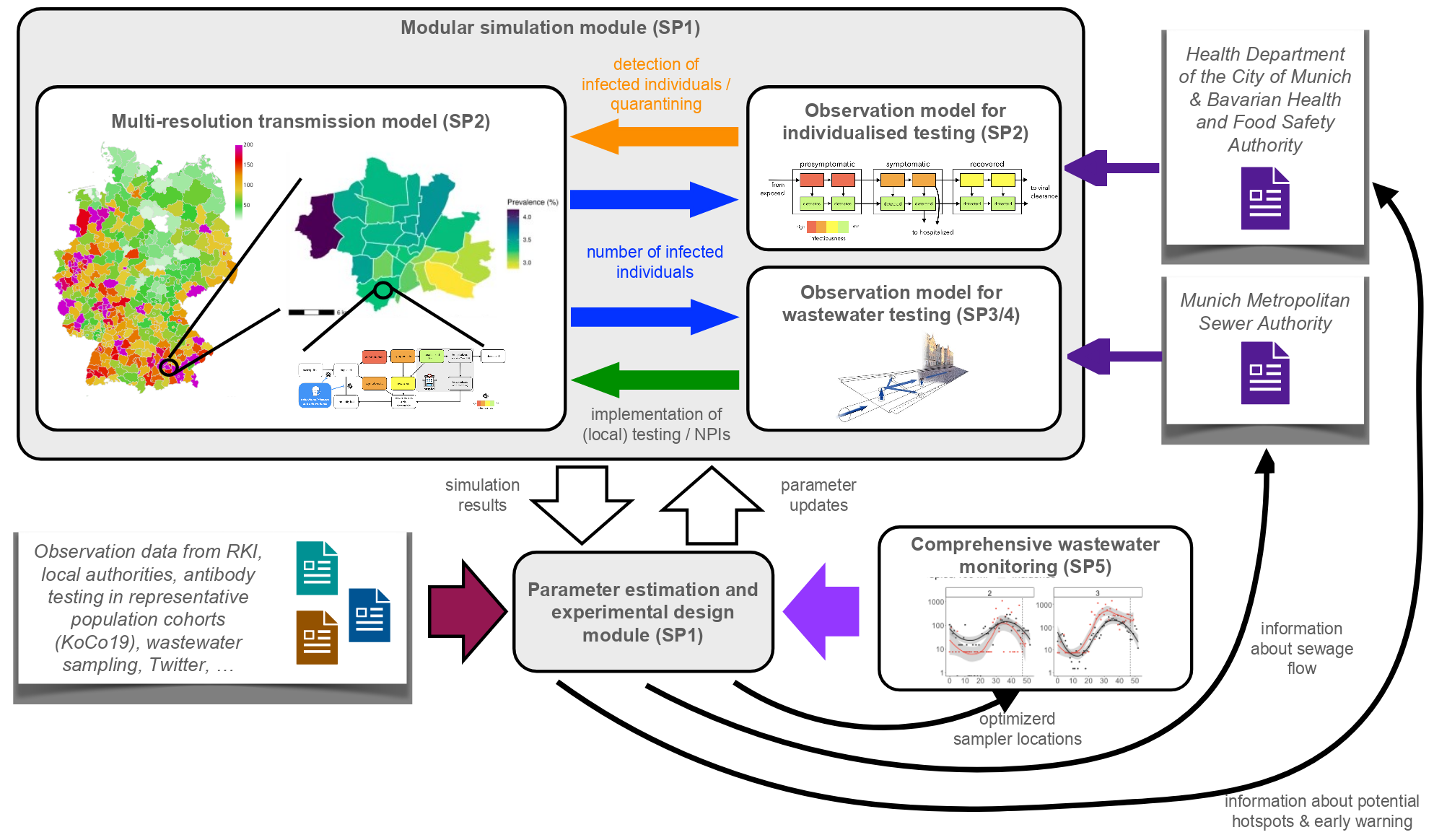
The technological basis of INSIDe are the three complementary state-of-the-art software frameworks pyABC [1], MEmilio [2, 3] and ++SYSTEMS [4], developed by the University of Bonn, the German Aerospace Center and Tandler respectively.
- pyABC enables data-driven modelling of multi-scale processes. In SP1, it is extended with ML methods to enable sequential updating of parameter estimates in longitudinal studies.
- MEmilio enables the simulation of the spatio-temporal spread of infectious diseases. In SP2, it will be extended with multiresolution simulation functionality to facilitate the study of processes occurring at different scales and equipped with models for the observational process.
- ++SYSTEMS enables the fine-mesh simulation of flow patterns in wastewater systems by the University of the Federal Armed Forces Munich (SP3). In SP4 it will be extended to enable the efficient creation of coarse-grained models for larger catchments.
We will use published and unpublished data for the parameterisation and validation of the improved models and the integrative pipeline. Most important is the unique access to data from the representative population and sanitation sample surveys in (i) Munich, Germany, and (ii) Addis Ababa and Jimma, Ethiopia, collected in projects coordinated by KUM. The available data will be supplemented by additional sample surveys (SP5).
The INSIDe platform will facilitate the assessment of the state of epidemics and pandemics, the analysis of their dynamics and the prediction of their further course. It will also enable data-driven evaluation of non-pharmaceutical interventions that benefit from a cross-disease and cross-strain approach. To this end, mechanistic descriptions of the interventions will be implemented in the model and effect sizes will be estimated using the combined information content of the different data sources mentioned above. The explicit description of the observation process (and thus the time-dependent probability of detecting an infected individual) will facilitate the deciphering of effects emanating from NPM (e.g. mask wearing, quarantine, venue closure, etc.) and changes in observation processes, e.g. test availability and strategy.
[1] Klinger E., Rickert D., Hasenauer J. pyABC: distributed, likelihood-free inference. bioRxiv. 2017; 162552.
[2] Kühn M.J., Abele D., Mitra T., Koslow W., Abedi M., Rack K., et al. MEmilio - a high performance Modular EpideMIcs simuLatIOn software. Available: https://github.com/DLR-SC/memilio.
[3] Kühn M.J., Abele D., Mitra T., Koslow W., Abedi M., Rack K., et al. Assessment of effective mitigation and prediction of the spread of SARS-CoV-2 in Germany using demographic information and spatial resolution. Math Biosci. 2021;339: 108648.
[4] tandler.com GmbH. ++Systems Isar. tandler.com GmbH; 18 Feb 2020 [cited 4 Oct 2021]. Available: https://tandler.com/de/page/software/plus-plus-systems.
